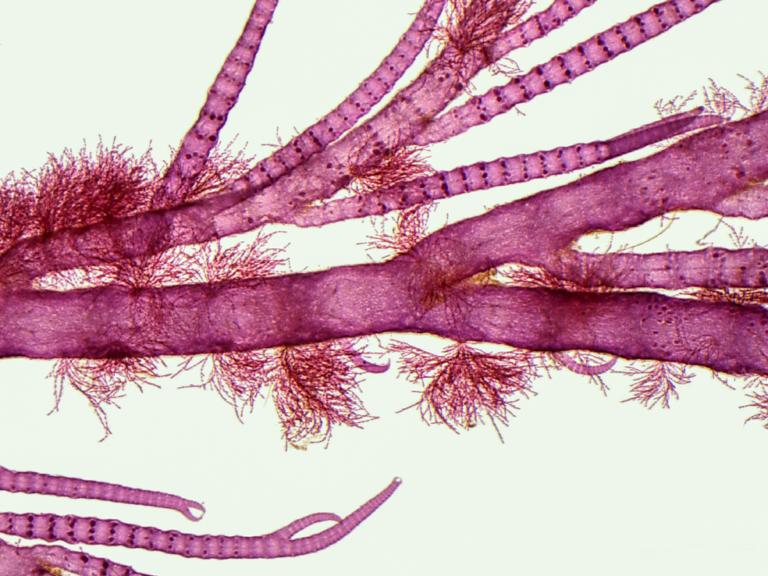
The current list of seaweed biodiversity in the Netherlands counts 300 species, yet little is known about their distribution and habitat preferences. This project investigates the taxonomic diversity and host specificity of epibiotic seaweeds, contributing to the Naturalis ARISE initiative.

Backgroundand context
Seaweeds are a diverse group of photosynthetic organisms found in coastal ecosystems worldwide. The current list of seaweed biodiversity in the Netherlands counts 300 species, but this number is likely far from complete. Many seaweed groups are challenging to distinguish based on morphology alone, which is especially problematic for the smaller, filamentous species. As a result, species diversity in these groups is probably much higher than currently recorded, and molecular data is essential for accurate identification. Additionally, little is known about their distribution and habitat preferences. These species often grow epibiotically on larger hosts, such as seaweeds and animals, but their specific host relationships remain unclear. This project investigates the taxonomic diversity and host specificity of epibiotic seaweeds, contributing to the Naturalis ARISE initiative, which aims to develop an infrastructure for identifying all Dutch species.
Objectivesand goals
The main goal of this project is to:
Assess taxonomic diversity and host-specificity of epibiotic seaweed species groups using molecular methods.
The student will develop the following skills:
- Sample collection during field work
- Morphological techniques for species identification (light microscopy, photography, making press-dried herbarium vouchers)
- Molecular techniques for DNA barcoding (DNA extraction, PCR, library preparation) with Sanger sequencing and Nanopore technologies
- Data analysis using Geneious and R
Materials and methods
Hosts that possibly harbour epibiotic seaweeds will be collected along the Dutch coast in different habitats and environments. This will involve work in the intertidal area and possibly snorkelling. In the lab, each host will be searched for the presence of small epibiotic seaweeds. These epibiotic species will then be morphologically characterized and photographed. All epibiotic seaweeds will be subsampled for DNA analysis. Different molecular protocols will be tested for sequencing with Sanger and Oxford Nanopore technologies to optimize sequencing of small seaweeds and mixed-species samples to capture the full biodiversity. Phylogenetic trees will be generated to assess species richness. Depending on the time availability of the student, it is also possible to morphologically compare the collected specimens to the historical collection available within Naturalis.
Studentrequirements
We are looking for a student who is enthusiastic about biodiversity of seaweeds. Knowledge of molecular lab work is preferred, but not essential.